Introduction
The SARS-CoV-2 virus has presented unprecedented challenges to healthcare systems worldwide, with severe cases frequently leading to acute respiratory distress syndrome (ARDS). A critical complication arising from ARDS and prolonged intensive care unit (ICU) stays is significant muscle wasting, also known as ICU-acquired weakness (ICUAW). This muscle loss can severely impact patient recovery and survival. While monitoring muscle mass is crucial in critical care settings, traditional methods have limitations. However, the increased use of computed tomography (CT) scans for COVID-19 patient management offers a unique opportunity to leverage body composition analysis (BCA) as an intermittent monitoring tool. This study investigates the application of an AI-powered “colored body tool” – referring to the visual segmentation of tissues in CT images – for critical care monitoring of muscle wasting in severe COVID-19 ARDS patients, and its correlation with patient outcomes.
Materials and Methods
Study Design and Patient Population
This retrospective cohort study analyzed data from critically ill patients with ARDS due to SARS-CoV-2 infection admitted to a university hospital ICU between March 2020 and January 2022. The study was approved by the Institutional Review Board and conducted in accordance with the Declaration of Helsinki. Inclusion criteria included: adult patients (>18 years), ARDS due to SARS-CoV-2, invasive mechanical ventilation (IMV) for ≥10 days, ICU stay ≥10 days, and availability of ≥3 serial CT scans including the abdomen. Out of 112 patients, 54 met the criteria, resulting in 239 CT examinations for analysis.
Body Composition Analysis using AI-Powered Colored Body Tool
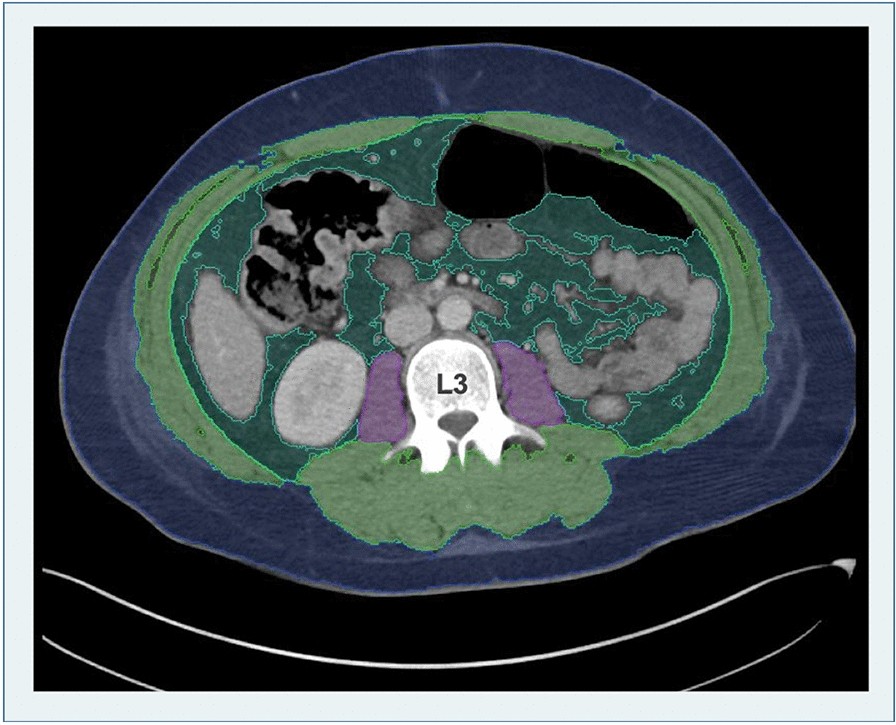
Body composition was assessed using an AI-based automated image segmentation tool integrated into the hospital’s Picture Archiving and Communication System (PACS) software (Visage version 7.1.). This “colored body tool” automatically identifies the third lumbar vertebra (L3) level and segments tissues in CT images into distinct colors representing subcutaneous fat (SAT), skeletal muscle area (SMA), visceral fat (VAT), and psoas muscle area (PMA). Figure 1 illustrates this colored segmentation. The software then quantifies these areas in square centimeters (cm²). Total abdominal muscle area (TAMA) was calculated as SMA + PMA. An experienced radiologist reviewed and corrected each automated segmentation to ensure accuracy.
Statistical Analysis
Statistical analysis was performed using Stata/MP version 16 and SPSS Statistics 27. Descriptive statistics, ANOVA, and linear mixed models were employed to analyze patient demographics, clinical characteristics, and body composition metrics. Relative psoas muscle loss per day was calculated for the entire monitoring period and between consecutive scans. Cox regression was used to identify associations with survival. Receiver operating characteristic (ROC) analysis and Youden index were used to determine a muscle loss cut-off for mortality prediction.
Results
Patient Demographics and Clinical Characteristics
The study cohort consisted of 54 critically ill patients (38 men, 16 women) with severe ARDS due to SARS-CoV-2. The mean age was 55.74 years, and the overall survival rate was 56.6%. Arterial hypertension was the most prevalent comorbidity (24/54 patients). Patients were mechanically ventilated for an average of 56 days and had a mean in-hospital stay of 80.2 days. Table 1 summarizes the clinical characteristics of the patient population.
Body Composition Changes and Muscle Loss
Initial BCA metrics from the first CT scans showed lower values in non-survivors, although not statistically significant. Linear mixed model analysis revealed a significant average loss per time point for skeletal muscle area (5.4 cm²) and psoas muscle area (1.3 cm²). Muscle distribution differed significantly between genders and ECMO therapy groups. PMA measurement proved to be the most reliable metric, less affected by fluid accumulation. The mean relative PMA loss between the first and last CT scan was 1.88% per day.
Non-survivors exhibited a significantly higher relative PMA decay per day (2.62%) compared to survivors (1.16%, p < 0.039). Figure 2 illustrates the PMA trends over time and examples of the “colored body tool” segmentation showing muscle loss. Maximum muscle decay was also significantly higher in men. Muscle loss between the first two CT scans did not significantly differ between survival groups, but showed a significant association with survival in Cox regression.
Pandemic Waves and Muscle Wasting
Analysis across three pandemic waves revealed the highest mortality and muscle loss rates in wave 2. The average daily psoas muscle loss was 3.58% in wave 2, compared to 2.97% in wave 1 and 1.72% in wave 3. Table 2 details the patient characteristics across different waves.
Survival Prediction and Discriminatory Cut-off
ROC analysis identified an overall PMA loss of 1.84% per day as a significant cut-off for survival prediction, with an AUC of 0.777 (Figure 3). Kaplan-Meier curves demonstrated significantly lower survival rates for patients exceeding this muscle loss threshold (p < 0.001).
Cox regression analysis revealed that lower initial TAMA and higher first PMA loss were significantly associated with decreased survival. Table 3 presents the Cox regression results.
Discussion
This study highlights the severity of muscle wasting in critically ill COVID-19 patients and its strong correlation with survival outcomes. The application of an AI-powered “colored body tool” for intermittent BCA using routinely acquired CT scans proved to be a valuable monitoring strategy in the critical care setting. The “colored body tool” allows for visual and quantitative assessment of muscle and fat compartments, providing critical insights into patient catabolism.
The observed mortality rate of 56.6% and prolonged hospitalization durations underscore the challenges in managing severe COVID-19. The study also corroborated the varying severity of COVID-19 waves, with wave 2 exhibiting the highest mortality and muscle loss.
Our findings align with previous research demonstrating rapid muscle decay in ICU patients and the prognostic value of muscle monitoring. This study extends these findings by examining long-term muscle wasting over the entire hospital stay and using clinically indicated CT scans for BCA. The intermittent nature of our monitoring, driven by clinical necessity rather than fixed intervals, reflects real-world critical care practice.
Compared to BIA and US, CT-derived BCA with the “colored body tool” offers more objective and reproducible measurements of multiple tissue components. The ability to use both in-house and externally acquired CT images enhances the feasibility of retrospective and long-term muscle atrophy analysis, especially in transferred patients. The increased availability of CT scans during the COVID-19 pandemic has made this approach particularly relevant and clinically applicable.
The identified PMA loss threshold of 1.84% per day and its strong discriminatory power for survival prediction suggest that BCA can contribute to risk stratification and potentially inform critical care decision-making, such as patient selection for ECMO therapy. Furthermore, monitoring muscle wasting with the “colored body tool” can aid in the early identification of patients at risk of ICUAW and PICS, enabling timely preventative and rehabilitative interventions.
Limitations
The retrospective design and moderate sample size are limitations. Selection bias may be present, as severely ill patients are overrepresented. Causality of muscle loss cannot be definitively established, and the impact of factors like NMBA and parenteral nutrition require further investigation. Generalizability may be limited by the specific patient population studied. Larger, prospective studies are needed to validate these findings and further explore the clinical utility of AI-powered BCA in critical care.
Conclusion
Severe muscle wasting is a significant complication in critically ill COVID-19 patients and is strongly associated with mortality. Intermittent BCA using an AI-powered “colored body tool” derived from clinically indicated CT scans offers a valuable monitoring tool in critical care. This method enables the identification of patients at high risk of adverse outcomes and holds substantial promise for supporting clinical decision-making and improving patient management in the ICU. The “colored body tool” approach transforms routine CT scans into a powerful resource for personalized critical care.
Acknowledgements
The authors thank Bettina Herwig and Camilla Pedersen for language editing.
Abbreviations
AI: Artificial intelligence
ANOVA: Analysis of variance
ARDS: Acute respiratory distress syndrome
BCA: Body composition analysis
BIA: Bioelectrical impendence analysis
BMI: Body mass index
CT: Computed tomography
ECMO: Extracorporeal membrane oxygenation
ICU: Intensive care unit
IMV: Invasive mechanical ventilation
NMBA: Neuromuscular blocking agents
PACS: Picture archiving and communication system
PICS: Post-intensive care syndrome
PMA: Psoas muscle area
pPeak: Peak inspiratory pressure
RMV: Respiratory minute volume
RR: Respiratory rate
SAT: Subcutaneous adipose tissue
SMA: Skeletal muscle area
SOFA: Sequential Organ Failure Assessment
sPEEP: Set positive end-expiratory pressure
TAMA: Total abdominal muscle area
US: Ultrasound
VAT: Visceral adipose tissue
Author contributions
JK, ZK, CP, and UF wrote the main text of the manuscript. Data curation was performed by JK, NLB, TA, TP and LS. Statistics and analysis were conducted by JK, UF and DG. All authors reviewed the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. Not applicable.
Availability of data and materials
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki and was approved by the Institutional Review Board (Internal registration number: EA4/152/20). Informed patient consent was waived by the IRB.
Consent for publication
Not applicable.
Competing interests
Uli Fehrenbach reports research agreements with Bayer Healthcare and fees for lectures (Bayer Healthcare, Guerbet, GE, Siemens, Ipsen) as well as for attending meetings (Bayer Healthcare, Asahi Intecc, Ipsen).
Tobias Penzkofer was supported by Berlin Institute of Health (Clinician Scientist Grant, Platform Grant), Ministry of Education and Research (BMBF, 01KX2021, 68GX21001A), German Research Foundation (DFG, SFB 1340/2), Horizon 2020 (952172) and reports research agreements (no personal payments, outside of submitted work) with AGO, Aprea AB, ARCAGY-GINECO, Astellas Pharma Global Inc. (APGD), Astra Zeneca, Clovis Oncology, Inc., Dohme Corp, Holaira, Incyte Corporation, Karyopharm, Lion Biotechnologies, Inc., MedImmune, Merck Sharp, Millennium Pharmaceuticals, Inc., Morphotec Inc., NovoCure Ltd., PharmaMar S.A. and PharmaMar USA, Inc., Roche, Siemens Healthineers, and TESARO Inc., and fees for a book translation (Elsevier).
All other authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Uli Fehrenbach and Dominik Geisel contributed equally to this work.
References
[References from the original article would be listed here]
Associated Data
Data Availability Statement
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.